Logistic Regression
This section covers the fundamental steps in the creation of
logistic regression models in the Logistic Regression
Platform of GeneXproTools. We’ll start with a quick hands-on introduction
to get you started, followed by a more detailed overview of the
fundamental tools you can explore in GeneXproTools to create
very good predictive models that accurately explain your data.
 |
Hands-on Introduction to Logistic Regression |
Designing a good logistic regression model in GeneXproTools is really simple:
after importing your data from Excel/Database or a
text file, GeneXproTools takes you immediately to the
Run Panel where you just have to click the
Start Button to create a model.
This is possible because GeneXproTools comes with pre-set default
parameters and data pre-processing procedures
(including dataset partitioning and handling
categorical variables and missing values) that work
very well with virtually all problems. We’ll learn later how to choose
some of the most basic settings so that you can explore all the advanced tools of
the application, but you can in fact quickly design highly sophisticated and accurate
logistic regression models in GeneXproTools with just a click.
Monitoring the Design Process
While the logistic regression model is being created by the learning algorithm, you can evaluate and
visualize the actual design process through the real-time
monitoring of different
model fitting charts and statistics in the Run Panel, such as
different Binomial Fitting Charts, the
Logistic Regression Scatter Plot, the ROC Curve, the
Logistic Regression Tapestry, the
Confusion Matrix,
the Logistic Regression Accuracy and the Fitness.
Model Evaluation & Testing
Then in the Results Panel you can further evaluate your model using different charts
and additional
measures of fit, such as the
area under the ROC curve, the sensitivity,
specificity, recall, precision and Matthews
correlation coefficient. It’s also in the Results Panel that you can check more thoroughly how your model
generalizes to unseen data by checking how well it performs in the
validation/test set.
Generating the Model Code
Then in the Model Panel you can see
and analyze the model code not only in the programming language of
your choice but also as a diagram representation or expression tree.
GeneXproTools includes 17 built-in programming
languages or grammars for Logistic Regression.
These grammars allow you to generate code automatically in
some of the most popular programming
languages around, namely Ada, C, C++, C#, Excel VBA, Fortran, Java, Javascript, Matlab,
Octave, Pascal, Perl, PHP, Python, R, Visual Basic,
and VB.Net.
But more importantly GeneXproTools also allows you
to add your own programming languages through
user-defined grammars, which can be easily
created using one of the existing grammars as
template.
Making Predictions
And finally, in the Scoring Panel of
GeneXproTools you can make predictions with your
models using
the generated JavaScript code. This means that you don’t have to know how to deploy the model code to
make predictions with your models outside GeneXproTools: you can make them
straightaway within the
GeneXproTools environment in the Scoring Panel.
GeneXproTools also
deploys automatically to Excel individual
models and
model
ensembles using the
generated Excel VBA code. More importantly,
GeneXproTools allows you to embed the Excel
VBA code of your models in the Excel worksheet, thus
allowing you to make predictions with your models very
conveniently in Excel. In addition, for model
ensembles, GeneXproTools also evaluates the
majority class model or the
average
probability model
and
median probability model, which are usually more robust and
accurate logistic regression models than individual models.
 |
Loading Data |
The logistic regression models
GeneXproTools creates are statistical in
nature or, in other words, data-based. Therefore
GeneXproTools needs training data
from which to extract the information needed to create the models. Data is also needed
for validating and testing the generalizability of the generated models.
However, validation/test data, although recommended,
is not mandatory and therefore need not be used if
data is in short supply.
Data Sources & Formats
Before evolving a model with GeneXproTools you must first
import the input data into GeneXproTools.
GeneXproTools allows you to import data from
Excel & databases,
text files and
GeneXproTools files.
Excel Files & Databases
The loading of data from Excel/databases requires making
a connection with Excel/database and then selecting the
worksheets or columns of interest. By default GeneXproTools will set
the last column as the
dependent or
response variable, but you can easily set any of the other columns
as response variable by selecting the variable of interest and then
checking Response in the context menu.
Text Files
For
text files GeneXproTools supports three
commonly used
data matrix formats.
The first is the standard
Records x Variables
(Response Last) format where records are
in rows and variables in columns, with the dependent or response variable occupying the
rightmost position. In the small example below with
seven records, iris_class is
the response variable and sepal_length, sepal_width, petal_length, and
petal_width are the independent or predictor variables:
|
sepal_length |
sepal_width |
petal_length |
petal_width |
iris_class |
|
4.9 |
3.1 |
1.5 |
0.1 |
0 |
|
6.7 |
3.3 |
5.7 |
2.5 |
1 |
|
5.1 |
3.5 |
1.4 |
0.2 |
0 |
|
5.8 |
2.6 |
4 |
1.2 |
0 |
|
4.3 |
3 |
1.1 |
0.1 |
0 |
|
5.7 |
2.9 |
4.2 |
1.3 |
0 |
|
5.6 |
2.8 |
4.9 |
2 |
1 |
The second format,
Records x Variables (Response First),
is similar to the first, with the difference that the response variable
is in the first column.
And the third data format is the
Gene Expression Matrix or GEM format commonly used
in DNA microarrays studies or whenever the number of variables far exceeds
the number of records. In this format records are in columns and
variables in rows, with the dependent variable
occupying the topmost position. For instance,
the small dataset above in GEM format maps to:
|
iris_class |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
|
sepal_length |
4.9 |
6.7 |
5.1 |
5.8 |
4.3 |
5.7 |
5.6 |
|
sepal_width |
3.1 |
3.3 |
3.5 |
2.6 |
3 |
2.9 |
2.8 |
|
petal_length |
1.5 |
5.7 |
1.4 |
4 |
1.1 |
4.2 |
4.9 |
|
petal_width |
0.1 |
2.5 |
0.2 |
1.2 |
0.1 |
1.3 |
2 |
This kind of format is the standard for
datasets with a relatively small number of records and thousands of
variables. Note, however, that this format is not
supported for Excel files and if your data is kept in this format in Excel, you must
copy it to a text file and then use this file to load your data into GeneXproTools.
GeneXproTools uses the Records x
Variables (Response Last) format internally and therefore all
kinds of input format are automatically
converted and shown in this format both in
the Data Panel and Scoring Panel.
For text files
GeneXproTools supports the standard
separators (space,
tab, comma, semicolon, and pipe) and detects them automatically. The
use of
labels to identify your variables is optional and
GeneXproTools also detects automatically whether they are
present or not. Note however that the use of labels
allows you to
generate more intelligible code where each variable
is clearly identified by
its name.
GeneXproTools Files
GeneXproTools files can be very convenient to use as
data source as they
allow the selection of exactly the same datasets
used in a particular run. This can be very useful especially if
you want to use the same datasets across different runs.
Loading Data Step-by-Step
To Load Input Data for Modeling
- Click the File Menu and then choose New.
The New Run Wizard appears. You must give a name to your new run file (the default filename extension of
GeneXproTools run files is .gep) and then choose
Logistic Regression in the Problem Category box and the kind of source file
in the Data Source Type box.
GeneXproTools allows you to work both with Excel
& databases, text
files and gep files.
- Then go to the Entire Dataset (or Training
Set) window by clicking the Next button.
Choose the path for the dataset by browsing the Open dialog
box and choose the appropriate data matrix format. Irrespective
of the data format used,
GeneXproTools shows the loaded data in the standard
Records x
Variables (Response Last) format, with the dependent variable occupying the
rightmost position.
- Then go to the Validation/Test Data window by clicking the Next button.
Repeat the same steps of the previous point if you wish to use a
specific validation/test set to evaluate the
generalizability of your models. The loading of
a specific validation/test set is optional as
GeneXproTools allows you to split your data into
different datasets for training and
testing/validation in the Dataset
Partitioning window.
- Click the Finish button to save your new run file.
The Save As dialog box appears and after choosing the directory where you want your new run file to be saved, the
GeneXproTools modeling environment appears.
Then you just have to click the Start button to create a model as
GeneXproTools automatically chooses from a gallery of templates default settings that will enable you to evolve a model
with just a click.
 |
Kinds of Data |
GeneXproTools supports both numerical and
categorical variables,
and for both types also supports
missing values. Categorical and
missing values are replaced automatically by simple
default mappings
so that you can create models straightaway, but you can choose
more appropriate mappings through the Category Mapping Window and
the Missing Values Mapping Window.
Numerical Data
GeneXproTools supports all kinds of tabular numerical datasets,
with variables usually in columns and records in rows. In GeneXproTools
all input data is converted to numerical data prior to model creation,
so numerical datasets are routine for GeneXproTools and sophisticated
visualization tools and statistical analyses are available in GeneXproTools
for analyzing these datasets.
As long as it fits in memory, the
dataset size is unlimited both for the training and validation/test
datasets. However, for big datasets efficient heuristics for splitting the
data are automatically applied when a run is created in order to ensure
an efficient evolution and good model generalizability. Note however that
these
default partitioning heuristics only apply if the data is loaded as a single file;
for data loaded using two different datasets you can access all the partitioning
(including the default) and sub-sampling schemes of GeneXproTools
in the
Dataset Partitioning Window and in the
General Settings Tab.
Categorical Data
GeneXproTools supports all kinds of
categorical variables, both as part of entirely
categorical datasets or intermixed with numerical variables. In
all cases, during data loading, the
categories in all categorical variables are automatically replaced by numerical values
so that you can start modeling straightaway.
GeneXproTools uses simple heuristics to make this initial mapping, but then lets you choose
more meaningful mappings in the
Category Mapping Window.
Dependent categorical variables are also supported in
Logistic Regression problems with more than two classes.
In these cases the
mapping is made so that only one class is singled out, resulting in a binomial outcome,
such as {0, 1} or {-1, 1}, which can then be used to create logistic regression models. The merging of the response variable in
Logistic Regression is handled in the
Class Merging & Discretization Window.
The beauty and power of GeneXproTools support for
categorical variables goes beyond
giving you access to a sophisticated and extremely useful tool for changing and
experimenting with different mappings easily and quickly by trying out different scenarios
and seeing immediately how they impact on modeling. Indeed GeneXproTools also generates
code
that supports data in exactly the same format that was loaded into GeneXproTools. This means
that all the code generated both for external model deployment or for scoring internally in
GeneXproTools, also supports categorical variables. Below is an example in
C++ of a
logistic regression model with both numeric and categorical variables.
//------------------------------------------------------------------------
// Logistic regression model generated by GeneXproTools 5.0 on 10/10/2013
// GEP File: D:\GeneXproTools\Version5.0\Tutorials\LoanRisk_02.gep
// Training Records: 667
// Validation Records: 333
// Fitness Function: Positive Correl, Logistic Threshold
// Training Fitness: 618.814006880155
// Training Accuracy: 77.51% (517)
// Validation Fitness: 598.428716314718
// Validation Accuracy: 77.18% (257)
//------------------------------------------------------------------------
#include "math.h"
#include "string.h"
double gepModel(char* d_string[]);
double gepMin2(double x, double y);
double gepMax2(double x, double y);
void TransformCategoricalInputs(char* input[], double output[]);
double gepModel(char* d_string[])
{
const double G2C2 = 2.37659895718253;
const double G4C1 = 4.83752952665792;
double d[20];
TransformCategoricalInputs(d_string, d);
double dblTemp = 0.0;
dblTemp = d[0];
dblTemp += (d[9]+(log(gepMin2((d[12]/d[10]),d[13]))+((d[8]+G2C2)/2.0)));
dblTemp += log((gepMax2(gepMin2((gepMin2(d[4],d[6])/(d[11]*d[11])),
gepMin2(d[16],d[13])),d[2])/d[1]));
dblTemp += log(pow(gepMax2(gepMin2(d[5],gepMin2((d[3]*d[17]),
(G4C1*d[15]))),d[19]),2));
const double SLOPE = 0.437452769716913;
const double INTERCEPT = -1.95464077161276;
double probabilityOne = 1.0 / (1.0 + exp(-(SLOPE * dblTemp + INTERCEPT)));
return probabilityOne;
}
double gepMin2(double x, double y)
{
double varTemp = x;
if (varTemp > y)
varTemp = y;
return varTemp;
}
double gepMax2(double x, double y)
{
double varTemp = x;
if (varTemp < y)
varTemp = y;
return varTemp;
}
void TransformCategoricalInputs(char* input[], double output[])
{
if(strcmp("A11", input[0]) == 0)
output[0] = 1.0;
else if(strcmp("A12", input[0]) == 0)
output[0] = 2.0;
else if(strcmp("A13", input[0]) == 0)
output[0] = 3.0;
else if(strcmp("A14", input[0]) == 0)
output[0] = 4.0;
else output[0] = 0.0;
output[1] = atof(input[1]);
if(strcmp("A30", input[2]) == 0)
output[2] = 1.0;
else if(strcmp("A31", input[2]) == 0)
output[2] = 2.0;
else if(strcmp("A32", input[2]) == 0)
output[2] = 3.0;
else if(strcmp("A33", input[2]) == 0)
output[2] = 4.0;
else if(strcmp("A34", input[2]) == 0)
output[2] = 5.0;
else output[2] = 0.0;
if(strcmp("A40", input[3]) == 0)
output[3] = 1.0;
else if(strcmp("A41", input[3]) == 0)
output[3] = 2.0;
else if(strcmp("A410", input[3]) == 0)
output[3] = 3.0;
else if(strcmp("A42", input[3]) == 0)
output[3] = 4.0;
else if(strcmp("A43", input[3]) == 0)
output[3] = 5.0;
else if(strcmp("A44", input[3]) == 0)
output[3] = 6.0;
else if(strcmp("A45", input[3]) == 0)
output[3] = 7.0;
else if(strcmp("A46", input[3]) == 0)
output[3] = 8.0;
else if(strcmp("A48", input[3]) == 0)
output[3] = 9.0;
else if(strcmp("A49", input[3]) == 0)
output[3] = 10.0;
else output[3] = 0.0;
output[4] = atof(input[4]);
if(strcmp("A61", input[5]) == 0)
output[5] = 1.0;
else if(strcmp("A62", input[5]) == 0)
output[5] = 2.0;
else if(strcmp("A63", input[5]) == 0)
output[5] = 3.0;
else if(strcmp("A64", input[5]) == 0)
output[5] = 4.0;
else if(strcmp("A65", input[5]) == 0)
output[5] = 5.0;
else output[5] = 0.0;
if(strcmp("A71", input[6]) == 0)
output[6] = 1.0;
else if(strcmp("A72", input[6]) == 0)
output[6] = 2.0;
else if(strcmp("A73", input[6]) == 0)
output[6] = 3.0;
else if(strcmp("A74", input[6]) == 0)
output[6] = 4.0;
else if(strcmp("A75", input[6]) == 0)
output[6] = 5.0;
else output[6] = 0.0;
if(strcmp("A91", input[8]) == 0)
output[8] = 1.0;
else if(strcmp("A92", input[8]) == 0)
output[8] = 2.0;
else if(strcmp("A93", input[8]) == 0)
output[8] = 3.0;
else if(strcmp("A94", input[8]) == 0)
output[8] = 4.0;
else output[8] = 0.0;
if(strcmp("A101", input[9]) == 0)
output[9] = 1.0;
else if(strcmp("A102", input[9]) == 0)
output[9] = 2.0;
else if(strcmp("A103", input[9]) == 0)
output[9] = 3.0;
else output[9] = 0.0;
output[10] = atof(input[10]);
if(strcmp("A121", input[11]) == 0)
output[11] = 1.0;
else if(strcmp("A122", input[11]) == 0)
output[11] = 2.0;
else if(strcmp("A123", input[11]) == 0)
output[11] = 3.0;
else if(strcmp("A124", input[11]) == 0)
output[11] = 4.0;
else output[11] = 0.0;
output[12] = atof(input[12]);
if(strcmp("A141", input[13]) == 0)
output[13] = 1.0;
else if(strcmp("A142", input[13]) == 0)
output[13] = 2.0;
else if(strcmp("A143", input[13]) == 0)
output[13] = 3.0;
else output[13] = 0.0;
output[15] = atof(input[15]);
if(strcmp("A171", input[16]) == 0)
output[16] = 1.0;
else if(strcmp("A172", input[16]) == 0)
output[16] = 2.0;
else if(strcmp("A173", input[16]) == 0)
output[16] = 3.0;
else if(strcmp("A174", input[16]) == 0)
output[16] = 4.0;
else output[16] = 0.0;
output[17] = atof(input[17]);
if(strcmp("A201", input[19]) == 0)
output[19] = 1.0;
else if(strcmp("A202", input[19]) == 0)
output[19] = 2.0;
else output[19] = 0.0;
}
Missing Values
GeneXproTools supports
missing values both for numerical and categorical variables.
The
supported representations for missing values consist of NULL, Null, null, NA, na, ?,
blank cells, ., ._, and .*, where * can be any letter in lower or upper case.
When data is loaded into GeneXproTools, the missing values are automatically replaced by
zero so that you can start modeling right away. But then GeneXproTools allows you to choose
different mappings through the
Missing Values Mapping Window.
In the Missing Values Mapping Window you have access to pre-computed
data statistics,
such as the majority class for categorical variables and the average for numerical variables,
to help you choose the most effective mapping.
As mentioned
above for categorical values, GeneXproTools is not just a useful platform
for trying out different mappings for missing values to see how they impact on
model evolution and then choose the best one: GeneXproTools generates
code with support
for missing values that you can immediately deploy without further hassle, allowing you
to use the exact same format that was used to load the data into GeneXproTools. The
sample
Matlab code below shows a
logistic regression model with missing values in
5 of the 16 input variables:
%------------------------------------------------------------------------
% Logistic regression model generated by GeneXproTools 5.0 on 10/10/2013
% GEP File: D:\GeneXproTools\Version5.0\Tutorials\CreditApproval_01.gep
% Training Records: 460
% Validation Records: 230
% Fitness Function: Positive Correl, Logistic Threshold
% Training Fitness: 842.341219982723
% Training Accuracy: 87.61% (403)
% Validation Fitness: 860.877550197044
% Validation Accuracy: 89.13% (205)
%------------------------------------------------------------------------
function result = gepModel(d_string)
G1C2 = 4.88082522049623;
G2C5 = 6.99087496566668;
G3C4 = -7.38278359324931;
G3C9 = -8.88058107242042;
G4C6 = 8.11926938688315;
G4C9 = 4.10061952574236;
d = TransformCategoricalInputs(d_string);
varTemp = 0.0;
varTemp = ((((d(6)+d(11))/2.0)+min(G1C2,d(2)))+(d(13)-d(3)));
varTemp = varTemp + (((G2C5-(gep3Rt(d(1))/(d(7)/d(13))))*d(9))^2);
varTemp = varTemp + ((d(9)*(((G3C4*G3C9)+(d(4)/G3C9))-((d(7)+d(10))+
d(7))))*d(10));
varTemp = varTemp + (((d(6)+((G4C6*d(3))-(G4C9-d(11))))/2.0)-(d(7)-
((d(11)+d(6))+d(7))));
SLOPE = 1.99869415155661E-02;
INTERCEPT = -6.06053816971452;
probabilityOne = 1.0 / (1.0 + exp(-(SLOPE * varTemp + INTERCEPT)));
result = probabilityOne;
function result = gep3Rt(x)
if (x < 0.0),
result = -((-x)^(1.0/3.0));
else
result = x^(1.0/3.0);
end
function output = TransformCategoricalInputs(input)
switch char(input(1))
case 'a'
output(1) = 1.0;
case 'b'
output(1) = 2.0;
case '?'
output(1) = 2.0;
otherwise
output(1) = 0.0;
end
switch char(input(2))
case '?'
output(2) = 0.0;
otherwise
output(2) = str2double(input(2));
end
output(3) = str2double(input(3));
switch char(input(4))
case 'l'
output(4) = 1.0;
case 'u'
output(4) = 2.0;
case 'y'
output(4) = 3.0;
case '?'
output(4) = 2.0;
otherwise
output(4) = 0.0;
end
switch char(input(6))
case 'aa'
output(6) = 1.0;
case 'c'
output(6) = 2.0;
case 'cc'
output(6) = 3.0;
case 'd'
output(6) = 4.0;
case 'e'
output(6) = 5.0;
case 'ff'
output(6) = 6.0;
case 'i'
output(6) = 7.0;
case 'j'
output(6) = 8.0;
case 'k'
output(6) = 9.0;
case 'm'
output(6) = 10.0;
case 'q'
output(6) = 11.0;
case 'r'
output(6) = 12.0;
case 'w'
output(6) = 13.0;
case 'x'
output(6) = 14.0;
case '?'
output(6) = 2.0;
otherwise
output(6) = 0.0;
end
switch char(input(7))
case 'bb'
output(7) = 1.0;
case 'dd'
output(7) = 2.0;
case 'ff'
output(7) = 3.0;
case 'h'
output(7) = 4.0;
case 'j'
output(7) = 5.0;
case 'n'
output(7) = 6.0;
case 'o'
output(7) = 7.0;
case 'v'
output(7) = 8.0;
case 'z'
output(7) = 9.0;
case '?'
output(7) = 8.0;
otherwise
output(7) = 0.0;
end
switch char(input(9))
case 'f'
output(9) = 1.0;
case 't'
output(9) = 2.0;
otherwise
output(9) = 0.0;
end
switch char(input(10))
case 'f'
output(10) = 1.0;
case 't'
output(10) = 2.0;
otherwise
output(10) = 0.0;
end
output(11) = str2double(input(11));
switch char(input(13))
case 'g'
output(13) = 1.0;
case 'p'
output(13) = 2.0;
case 's'
output(13) = 3.0;
otherwise
output(13) = 0.0;
end
Normalized Data
GeneXproTools supports different kinds of
data normalization (
Standardization,
0/1 Normalization and
Min/Max Normalization), normalizing all numeric input variables
using
data statistics derived from the training dataset. This means that the validation/test dataset
is also normalized using the
training data statistics such as averages, standard deviations,
and min and max values evaluated for all numeric variables.
Data normalization can be beneficial for datasets with variables in very different
scales or ranges. Note, however, that data normalization is not a requirement even
in these cases, as the learning algorithms of GeneXproTools can handle
unscaled data
quite well. But since it might help, GeneXproTools allows you to see very quickly and
easily if normalizing your data improves modeling: if not, also as quickly,
you can revert to the original raw data.
It’s worth pointing out that GeneXproTools offers not just a convenient way of trying out
different normalization schemes. As is the case for
categorical variables and
missing values,
GeneXproTools generates
code that also supports data scaling, allowing you to deploy
your models confidently knowing that you can use exactly the same data format that
was used to load the data into GeneXproTools. Below is a sample code in
R of a
logistic regression model created using data standardized in the GeneXproTools environment.
#------------------------------------------------------------------------
# Logistic regression model generated by GeneXproTools 5.0 on 10/10/2013
# GEP File: D:\GeneXproTools\Version5.0\Tutorials\CreditApproval_02.gep
# Training Records: 460
# Validation Records: 230
# Fitness Function: Positive Correl, Logistic Threshold
# Training Fitness: 835.150160022832
# Training Accuracy: 86.52% (398)
# Validation Fitness: 849.303057612163
# Validation Accuracy: 87.83% (202)
#------------------------------------------------------------------------
gepModel <- function(d_string)
{
G1C3 <- 8.68892483291116
G3C9 <- 2.25501266518143
d <- TransformCategoricalInputs(d_string)
d <- Standardize(d)
dblTemp <- 0.0
dblTemp <- (((gep3Rt(d[4])*min(d[7],d[11]))*d[12])+((d[9]+G1C3) ^ 2))
dblTemp <- dblTemp + min(d[13],((d[7]+(d[5]-d[9])) ^ 2))
dblTemp <- dblTemp + (min(((d[15]+d[1])-(d[2]*d[3])),d[15])-(d[13]*(G3C9 ^ 2)))
dblTemp <- dblTemp + (log(max((d[8]+(d[10]+(d[13]+d[14]))),d[6]))*d[3])
SLOPE <- 0.166665156925704
INTERCEPT <- -16.6420098610091
probabilityOne <- 1.0 / (1.0 + exp(-(SLOPE * dblTemp + INTERCEPT)))
return (probabilityOne)
}
gep3Rt <- function(x)
{
return (if (x < 0.0) (-((-x) ^ (1.0/3.0))) else (x ^ (1.0/3.0)))
}
TransformCategoricalInputs <- function (input)
{
output <- rep(0.0, 15)
output[1] = switch (input[1],
"a" = 1.0,
"b" = 2.0,
"?" = 2.0,
0.0)
output[2] = switch (input[2],
"?" = 0.0,
as.numeric(input[2]))
output[3] = as.numeric(input[3])
output[4] = switch (input[4],
"l" = 1.0,
"u" = 2.0,
"y" = 3.0,
"?" = 2.0,
0.0)
output[5] = switch (input[5],
"g" = 2.0,
"gg" = 1.0,
"p" = 3.0,
"?" = 2.0,
0.0)
output[6] = switch (input[6],
"aa" = 1.0,
"c" = 2.0,
"cc" = 3.0,
"d" = 4.0,
"e" = 5.0,
"ff" = 6.0,
"i" = 7.0,
"j" = 8.0,
"k" = 9.0,
"m" = 10.0,
"q" = 11.0,
"r" = 12.0,
"w" = 13.0,
"x" = 14.0,
"?" = 2.0,
0.0)
output[7] = switch (input[7],
"bb" = 1.0,
"dd" = 2.0,
"ff" = 3.0,
"h" = 4.0,
"j" = 5.0,
"n" = 6.0,
"o" = 7.0,
"v" = 8.0,
"z" = 9.0,
"?" = 8.0,
0.0)
output[8] = as.numeric(input[8])
output[9] = switch (input[9],
"f" = 1.0,
"t" = 2.0,
0.0)
output[10] = switch (input[10],
"f" = 1.0,
"t" = 2.0,
0.0)
output[11] = as.numeric(input[11])
output[12] = switch (input[12],
"f" = 1.0,
"t" = 2.0,
0.0)
output[13] = switch (input[13],
"g" = 1.0,
"p" = 2.0,
"s" = 3.0,
0.0)
output[14] = switch (input[14],
"?" = 0.0,
as.numeric(input[14]))
output[15] = as.numeric(input[15])
return (output)
}
Standardize <- function (input)
{
AVERAGE_2 = 31.2013695652174
STDEV_2 = 12.4979385522111
input[2] = (input[2] - AVERAGE_2) / STDEV_2
AVERAGE_3 = 4.74421739130435
STDEV_3 = 4.84263178556765
input[3] = (input[3] - AVERAGE_3) / STDEV_3
AVERAGE_8 = 2.18686956521739
STDEV_8 = 3.02606706802097
input[8] = (input[8] - AVERAGE_8) / STDEV_8
AVERAGE_11 = 2.21739130434783
STDEV_11 = 3.88625331874295
input[11] = (input[11] - AVERAGE_11) / STDEV_11
AVERAGE_14 = 177.708695652174
STDEV_14 = 155.270169808247
input[14] = (input[14] - AVERAGE_14) / STDEV_14
AVERAGE_15 = 1077.55652173913
STDEV_15 = 5812.28518492274
input[15] = (input[15] - AVERAGE_15) / STDEV_15
return (input)
}
 |
Datasets |
Through the
Dataset Partitioning Window and the
sub-sampling schemes in the General Settings Tab,
GeneXproTools allows you to split your data into different datasets that can be used to:
- Create the models (the training dataset or a sub-set of the training dataset).
- Check and select the models during the design process (the validation dataset or a sub-set of the validation dataset).
- Test the final model (a sub-set of the validation set reserved for testing).
Of all these datasets, the
training dataset is the only one that is mandatory as GeneXproTools
requires data to create data models. The
validation and
test sets are optional and
you can indeed
create models without checking or testing them. Note however that this approach is not recommended
and you have indeed better chances of creating good models if you check their
generalizability
regularly not only
during
model design but also during
model selection. However if you don’t have
enough data,
you can still create good models with GeneXproTools as the
learning algorithms of GeneXproTools
are not prone to
overfitting the data. In addition, if you are using GeneXproTools to create
random forests, the need for validating/testing the models of the ensemble is less important
as ensembles tend to generalize better than individual models.
Training Dataset
The
training dataset is used to create the models, either in its entirety or as a
sub-sample
of the training data. The sub-samples of the training data are managed in the Settings Panel.
GeneXproTools supports different
sub-sampling schemes, such as
bagging and
mini-batch.
For example, to operate in
bagging mode you just have to set the sub-sampling to
Random.
In addition, you can also change the number of records used in each bag, allowing you
to speed up evolution if you have enough data to get good generalization.
Besides
Random Sampling (which is done with replacement) you can also choose
Shuffled
(which is done without replacement),
Balanced Random (in which a sub-sample is
randomly generated so that the proportion of
positives and negatives are the same),
Balanced Shuffled
(similar to Balanced Random,
but with the sampling done without replacement),
Odd/Even
Cases (particularly useful for time series
data), and different
Top/Bottom partition
schemes with control over the number of records
drawn either from the top or bottom of the dataset.
All types of random sampling can be used in
mini-batch mode, which is an extremely useful
sampling method for handling big datasets. In mini-batch mode a sub-sampling of the
training data is generated each
p generations (the
period of the mini-batch, which
is adjustable and can be set in the Settings Panel) and is used for training
during that period. This way,
for large datasets good models can be generated quickly using an
overall high percentage of
the records in the training data, without stalling the whole evolutionary process
with a huge dataset that is used each generation. It’s important however to find a
good balance between the size of the mini-batch and an efficient model evolution.
This means that you’ll still have to choose an appropriate number of records in order to ensure
good generalizability, which is true for all datasets, big and small. A simple rule
of thumb is to see if the best fitness is increasing overall: if you see it fluctuating
up and down, evolution has stalled and you need either to increase the mini-batch size or
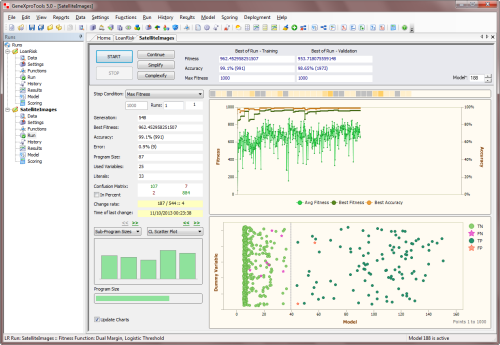
increase the time between batches (the period). The chart below shows clearly the overall
upward trend in best fitness for a run in mini-batch mode with a period of
50.
The training dataset is also used for evaluating
data statistics that are used in certain models,
such as the average and the standard deviation of predictor variables used in models created
with
standardized data. Other data statistics used in GeneXproTools include: pre-computed
suggested mappings for
missing values; training data constants used in Excel worksheets
both for models and ensembles deployed to Excel; min and max values of variables when
normalized data
is used (
0/1 Normalization and
Min/Max
Normalization), and so on.
It’s important to note that when a sub-set of the training dataset is used in a run, the
training data constants pertain to the data constants of the entire training dataset as
defined in the
Data Panel. For example, this is important when designing
ensemble models
using different random sampling schemes selected in the Settings Panel.
Validation Dataset
GeneXproTools supports the use of a
validation dataset, which can be either loaded as a
separate dataset or generated from a single dataset using GeneXproTools
partitioning algorithms.
Indeed if during the creation of a new run a single dataset is loaded, GeneXproTools automatically
splits the data into Training and Validation/Test datasets. GeneXproTools uses optimal strategies
to split the data in order to ensure good model design and evolution. These
default partition strategies
offer useful guidelines, but you can choose different partitions in the
Dataset Partitioning Window
to meet your needs. For instance, the
Odds/Evens partition is useful for
times series data, allowing
for a good split without losing the time dimension of the original data, which obviously can help in
better understanding both the data and the generated models.
GeneXproTools also supports
sub-sampling for the validation dataset, which, as explained for the
training dataset
above, is controlled in the Settings Panel.
The sub-sampling schemes available for the validation data are exactly the same available for the
training data, except of course for the
mini-batch strategy which pertains only to the training data.
It’s worth pointing out, however, that the same sampling scheme in the training and validation data
can play very different roles. For example, by choosing the Odds or the Evens, or the Bottom Half or
Top Half for validation, you can reserve the other part for testing
and only use this
test dataset at the very
end of the modeling process to evaluate the accuracy
of your model.
Another ingenious use of the random sampling schemes available for
the validation set in the Logistic Regression Platform (especially Random
and
Shuffled sub-sampling) consists of calculating the
cross-validation accuracy of
a model. A clever and simple way to do this, consists of creating a run with the model you want
to cross-validate. Then you can copy this model
n times, for instance by importing it
n times.
Then in the History Panel you can evaluate the performance of the model for different sub-samples
of the validation dataset. The average value for the fitness and
favorite statistic shown in the
statistics summary of the History Panel consist of the cross-validation results for your model.
Below is an example of a 30-fold cross-validation
evaluated for the training and validation datasets
using random sampling of the respective datasets.
Test Dataset
The dividing line between a
test dataset and a
validation dataset is not always clear.
A popular
definition comes from modeling competitions, where part of the data is hold out and not
accessible to the
people doing the modeling. In this case, of course, there’s no other choice: you create your
model and then others check if it is any good or not. But in most real situations people do have
access to all the data and they are the ones who decide what goes into training, validation and
testing.
GeneXproTools allows you to experiment with all these scenarios and you can choose what works best
for the data and problem you are modeling. So if you want to be strict, you can hold out part of
the data for testing and load it only at the very end of the modeling process, using the
Change Validation Dataset functionality of GeneXproTools.
Another option is to use the technique described
above for the validation dataset, where you
hold out part of the validation data for testing. For example, you hold
out the Odds or the Evens,
or the Top Half
or Bottom Half. This obviously requires strong willed and very disciplined people, so it’s perhaps
best practiced only if a single person is doing the modeling.
The takeaway message of all these what-if scenarios is that, after working with GeneXproTools
for a while, you’ll be comfortable with what is good practice in testing the accuracy of
the models you create with it. We like to claim that
the learning algorithms of GeneXproTools are not prone to
overfitting and now
with all the partitioning and sampling schemes of GeneXproTools you can develop a better sense of
the quality of the models generated by GeneXproTools.
And finally, the same
cross-validation technique described above
for the validation dataset can be
performed for the test dataset.
 |
Choosing the Function Set |
GeneXproTools allows you to choose your function set from a total of
279 built-in mathematical functions and an unlimited number of
custom functions, designed using the JavaScript language in the GeneXproTools environment.
Built-in Mathematical Functions
GeneXproTools offers a total of 279 built-in mathematical
functions, including 186 different if then else rules, that
can be used to design both linear and nonlinear logistic regression models. This wide
range of mathematical functions allows the evolution of
highly sophisticated
and accurate models, easily built with the most appropriate functions.
You can find the description of all the 279 built-in mathematical functions
available in GeneXproTools, including their representation
here.
The Function Selection Tools of GeneXproTools
can help you
in the selection of different function sets very quickly
through the combination of the Show options with the
Random, Default, Clear, and Select All buttons plus the
Add/Reduce
Weight buttons in the Functions
Panel.
User Defined Functions
Despite the great diversity of GeneXproTools
built-in mathematical functions, some users
sometimes want to model with different ones.
GeneXproTools gives the user the possibility
of creating custom functions (called
Dynamic UDFs and represented as DDFs
in the generated code) in order to evolve models with them.
Note however that the use of custom functions is
computationally demanding, slowing considerably the evolutionary process and therefore should be used with moderation.
By selecting the Functions Tab in the Functions Panel, you have full access to
all the available functions, including all the
functions you've designed and all the built-in math functions. It's also here in the Functions Panel
that you add
the custom functions (Dynamic UDFs or DDFs) to your modeling
toolbox.
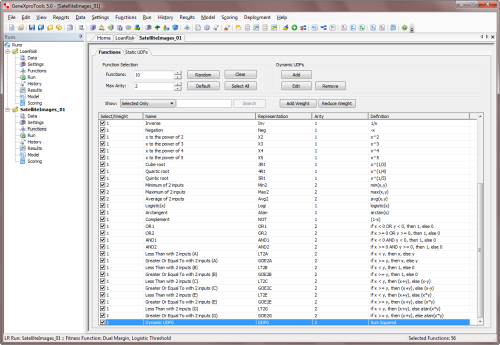
To add a custom function to your function set, just check the checkbox on the Select/Weight column and
select the appropriate weight for the function (the weight determines the probability of each function being drawn
during mutation and other random events in the creation/modification of programs).
By default, the weight of each newly added function is 1, but you can increase the probability of a function being included in your models by increasing its weight in the Select/Weight column. GeneXproTools automatically balances your
function set with the number of independent variables in your data,
therefore you just have to select the set of functions for your problem and then choose their relative
proportions by choosing their weights.
To create a new custom function, just click the
Add button on the Dynamic UDFs frame and the
DDF Editor appears. You can also edit old functions
through the
Edit button or remove them
altogether from your modeling toolbox by clicking
the
Remove button.
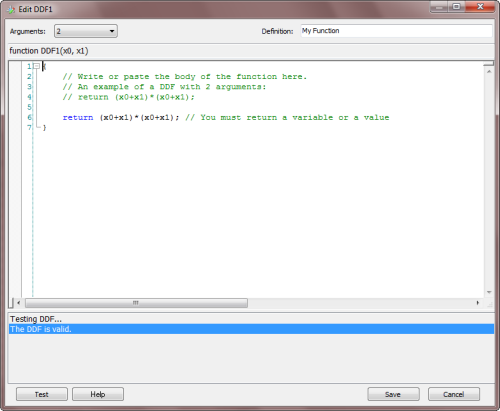
By choosing the number of arguments (minimum is 1 and maximum is 4) in the
Arguments combobox, the function header appears
in the code window. Then you just have to write the body of the function in the code editor. The code must be in JavaScript and can be
conveniently tested for compiling errors by clicking the Test button.
In the Definition box, you can write a brief description of the function for your future reference. The text you write
there will appear in the Definition column in the Functions Panel.
Dynamic UDFs are extremely powerful and interesting tools as they are treated exactly
like the built-in functions of GeneXproTools and therefore can be used to model
all kinds of relationships not only between the original variables but also between
derived features created on the fly by the learning algorithm. For instance, you can design
a DDF so that it will model the log of the sum of four expressions, that is,
DDF = log((expression 1) + (expression 2) + (expression 3) + (expression 4)),
where the value of each expression will depend on the context of the DDF in the
program.
 |
Creating Derived Features/Variables |
Derived variables or
new features can be easily created in GeneXproTools
from the original variables.
They are created in the Functions Panel, in the Static UDFs Tab.
Derived variables were originally called
UDFs or
User Defined Functions
and therefore in the code generated by GeneXproTools they are represented as UDF0, UDF1, UDF2, and so on. Note however that
UDFs are in fact new features derived from the original variables in the training and
validation/test datasets.
Like
DDFs, they are implemented in JavaScript using the JavaScript editor of GeneXproTools.
These user defined features are then used by the learning algorithm exactly as
the original features, that is, they are incorporated into the evolving models
adaptively, with the most important being chosen and selected according to
how much they contribute to the performance of each
model.
 |
Choosing the Model Architecture |
In GeneXproTools the candidate solutions or models are
encoded in linear strings or chromosomes with
a special architecture. This architecture includes
genes with different gene domains (head,
tail, and
random constants domains) and a linking function to
link all the genes. So the parameters that you can
adjust include the
head size, the number of genes and the linking
function. You set the values for these parameters in the Settings Panel -> General
Settings Tab.
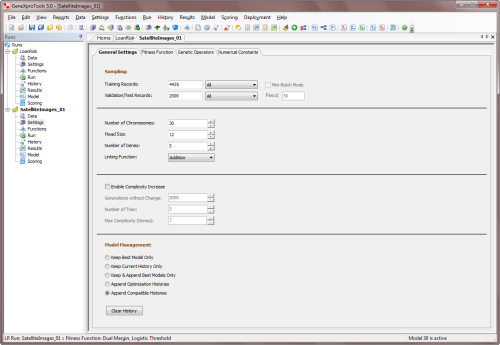
The Head Size determines the complexity or
maximum size of each term in your model. In the
heads of genes, the learning algorithm tries out different arrangements of functions and
terminals (original & derived variables and constants) in order to model your data.
The plasticity of this architecture allows the creation of an infinite number of models
of different sizes and shapes. A small number of these models (a
population) is
randomly generated and then tested to see how well
each model explains the data. Then according
to their performance or fitness the models are selected to reproduce with some minor changes,
giving rise to new models. This process of selection and
reproduction is repeated for
a certain number of generations, leading to the discovery of better and better models.
The heads of genes are shown in blue in the compact
linear representation (Karva notation) of the model
code in the Model Panel.
This linear code, which is the representation
that the learning algorithms of GeneXproTools use
internally, is then translated into any
of the built-in programming languages of
GeneXproTools (Ada, C, C++, C#, Excel VBA, Fortran, Java, JavaScript,
Matlab, Octave, Pascal, Perl, PHP, Python, R, Visual Basic, and VB.Net)
or any other language you add through the use of
GeneXproTools custom
grammars.
More specifically, the head size h of each gene determines the maximum width
w and maximum depth d of the sub-expression trees
encoded in each gene, which are given by the formulas:
w = (n - 1) *
h + 1
d = ((h + 1) /
m) * ((m +
1) / 2)
where m is minimum arity (the smallest
number of arguments taken by the functions in the
function set) and n is maximum arity (the
largest number of arguments taken by the functions
in the function set).
GeneXproTools also allows you to visualize clearly the model structure
and composition by showing the expression trees of all your models in the Model Panel.
Thus, the learning algorithm selects its
models between the minimum possible size (which is a
model composed only of one-element trees) and the
maximum allowed size,
fine-tuning the ideal size and shape during
the evolutionary process, creating and
testing new features on the fly
without human intervention.
The
number of genes per chromosome is also an important parameter. It determines the number of
fundamental
terms or
building blocks in your models as each gene codes for a different
sub-expression tree (sub-ET). Theoretically, one could just use a huge single gene
in order to evolve very complex models. However, by
dividing the chromosome into simpler,
more manageable units gives an edge to the learning process and more efficient and elegant models
can be discovered this way.
Whenever the number of genes is greater than one, you must also choose a suitable
linking function to connect the mathematical terms encoded in each gene.
GeneXproTools allows you to choose
addition,
subtraction,
multiplication,
division,
average,
min, and
max to link the sub-ETs. As expected, addition, subtraction
and average work very well for virtually all problems but sometimes one of the other linkers could be useful for searching different solution spaces.
For example, if you suspect the solution to a problem involves a quotient between two big terms, you can choose two genes linked by division.
 |
Choosing the Fitness Function |
For Logistic Regression
problems, in the Fitness
Function Tab of the
Settings Panel you have
access to a a total of
59
built-in fitness
functions, most of which combine multiple objectives, such as the use
of different reference simple models, cost matrix,
different types of rounding thresholds, including evolvable thresholds,
lower and upper bounds for
the model output, parsimony pressure, variable pressure, and many more.
Additionally, you can also design your own
custom fitness functions and explore the solution space with
them.
By clicking the Edit Custom Fitness button, the
Custom
Fitness
Editor is opened and there you can write the code of
your fitness function in JavaScript.
The kind of fitness function you choose will depend most probably on the
cost function
or error measure you are most familiar with. And although there is nothing wrong with this,
for all of them can accomplish an efficient
evolution, you might want to try different
fitness functions for they travel the
fitness landscape differently: some of them
very straightforwardly in their pursuits
while others choose less travelled paths,
considerably enhancing the search process. Having
different fitness functions in your modeling toolbox is also
essential in the design of ensemble models.
 |
Exploring the Learning Algorithms |
GeneXproTools uses two different learning algorithms for
Logistic Regression problems. The first – the basic gene expression algorithm
or simply
Gene Expression Programming (GEP) – does not support the direct manipulation of random numerical constants,
whereas the second – GEP with Random Numerical Constants or
GEP-RNC
for short – implements a structure for handling them directly. These
two algorithms search the solution landscape differently and
therefore it might be a good idea to try them both on your problems.
For example, GEP-RNC models are usually more
compact than models generated without random
numerical constants.
The kinds of models these algorithms produce are quite different
and, even if both of them perform equally well on the problem at hand,
you might still prefer one over the other. But there are cases, however,
where numerical constants are crucial for an efficient modeling and,
therefore, the GEP-RNC algorithm is the default in
GeneXproTools. You activate this algorithm in the Settings Panel ->
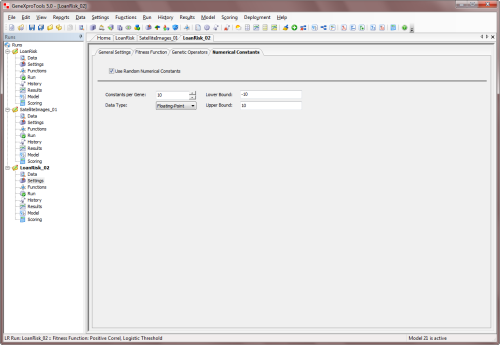
Numerical Constants by checking the Use Random Numerical Constants checkbox.
In the Numerical Constants tab you can also adjust the range and
type of constants and also the number of constants per gene.
The GEP-RNC algorithm is slightly more complex than
the basic gene expression algorithm as it uses an additional gene domain (Dc) for encoding the random
numerical constants. Consequently, this algorithm
includes an additional set of genetic operators (RNC
Mutation, Constant Fine-Tuning, Constant Range
Finding, Constant Insertion, Dc Mutation, Dc Inversion, Dc IS
Transposition, and Dc Permutation) especially developed for handling random
numerical constants (if you are not familiar with these operators,
please use the default Optimal Evolution Strategy by
selecting Optimal Evolution in the Strategy combobox
as it works very well in all cases; or you can learn more about the
genetic operators in
the
Legacy Knowledge Base).
 |
Exploring Different Evolutionary Strategies for an Efficient Learning |
Predicting unknown behavior efficiently is of course the foremost goal in modeling.
But extracting knowledge from the blindly designed models is
also extremely important
as this knowledge can be used not only to enlighten further the modeling process but also
to understand the complex relationships between variables.
So, the evolutionary strategies we recommend in the GeneXproTools
templates
for Logistic Regression reflect these two main concerns: efficiency and simplicity. Basically, we recommend starting the modeling process with the
GEP-RNC algorithm
and a function set well adjusted to the complexity of the problem.
GeneXproTools selects the appropriate template for your problem according
to the number of independent variables in your data. This kind of template is a good starting
point that allows you to start the modeling process
straightaway with just a mouse click. Indeed, even if you are not familiar with evolutionary computation in general and
Gene Expression Programming in particular, you will be able to design complex logistic regression models
(both linear and nonlinear) immediately thanks to the templates of
GeneXproTools. In these templates, all the adjustable parameters of the
default learning algorithm are already set
and therefore you don’t have to know how to create genetic diversity, how to set the appropriate population size, the chromosome architecture,
the number of constants, the type and range for the
random constants, the fitness function, the stop
condition, and so forth. Then, as you learn more about
GeneXproTools, you will be able to explore all its modeling tools and create quickly and efficiently very good
logistic regression models that will allow you to understand
and model your data like never before.
So, after creating a new run you just have to click the
Start button in the
Run Panel in order to design a logistic regression model.
GeneXproTools allows you
to monitor the evolutionary process by giving you
access to
different model fitting charts, including different
curve fitting charts, scatter plots and residual
plots. Then, whenever you see fit, you can stop the run
(by clicking the
Stop button) without fear of stopping
evolution prematurely as
GeneXproTools allows you to continue to improve the
model by using the best model thus far (or any other
model) as the starting
point (
evolve with seed
method). For that you just have to click the
Continue button in the
Run Panel to continue the search for a
better model. Alternatively you can simplify or
complexify your model by clicking either the
Simplify or
Complexify buttons.
This strategy has enormous advantages as you might choose to stop the run at any time and then take a closer look at the evolved model. For instance, you can analyze its mathematical representation, its performance in the
validation or test set,
evaluate essential statistics and measures of fit
for a quick and rigorous assessment of its accuracy,
see how it performs on a different test set, and so on. Then you might choose to adjust a few parameters, say, choose a different fitness function, expand the function set, add a neutral gene,
apply parsimony pressure for simplifying its
structure, change the training set for model
refreshing, and so on, and then explore this new
set of conditions to further improve the model. You can repeat this process for as long as you want or until you are completely satisfied with
your model.

