 |
Introduction |
A job definition is an XML file which contains processing instructions to be applied to a template gep file. The complete process requires a job file and a gep file as a template. The instructions in the job file will be applied to copies of the gep file which are created when the job file is processed by GeneXproServer. Here is an example of a very simple job file:
<job filename='CreditApproval.gep' path='c:\work' feedback='2'
usesubfolder='1' async='yes'>
<run id='1' stopcondition='Minutes' value='30'/>
<run id='2' stopcondition='Minutes' value='45'/>
</job>
The initial state of the folder would be:
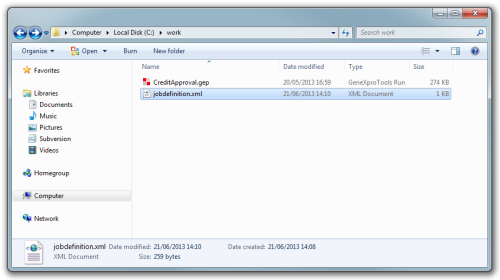
This job file will use the CreditApproval.gep file which is in the folder c:\work as template. It will process in parallel (async='yes') two runs, one for 30 minutes and the second one for 45 minutes. Each one of the runs will be a copy of the original CreditApproval.gep file and will be placed in one of two sub-folders named CreditApproval_1 and CreditApproval_2. By default GeneXproServer will generate a report for each new run that includes logging and summary information.
To start a run use the command (assuming that you chose to add GeneXproServer
to the PATH during installation):
gxps50c.exe c:\work\jobdefinition.xml
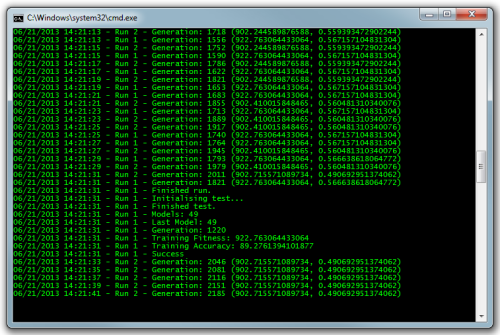
The following image shows the transition between processing two runs simultaneously and finishing the second run which will run for longer:
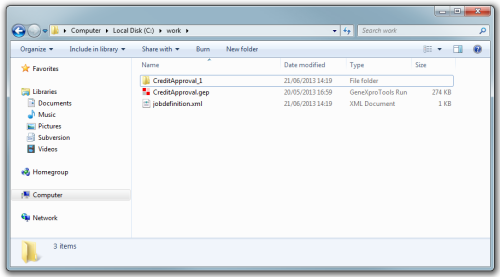
Finally when all processing is done the contents of the folder c:\work will be:
And inside the CreditApproval_1 you will find the following files:
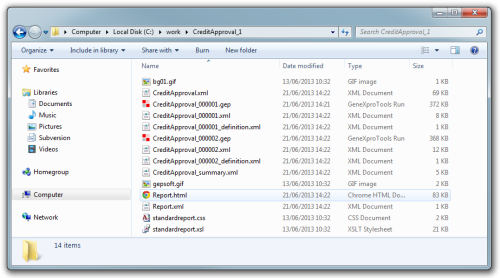
These include the two new gep files (CreditApproval_000001.gep and CreditApproval_000002.gep), several report files for each run and a summary report in xml and html (Report.xml and Report.htm).
This describes a simple job but there are many options that can be added to the process such as running external processes before and after the job and before and after each run, code generation, testing,
and so on. These options are detailed below.
 |
Job Node |
In a job definition file the job node is the top node and encloses all the commands in the file. The job itself supports the following attributes:
Required Attributes:
filename: The name of the gep file including the extension.
path: The fully qualified path to the folder where the gep file is. All processing will be done in this folder.
feedback: The amount of time in seconds between updates to the screen.
Optional Attributes:
usesubfolder: Create a folder for the job. When set to 1 (Auto) the folder will be called filename_x where
x is an integer starting at 1. If set to 2 (Fixed) then the attribute
subfoldername is required and must contain a valid file name.
Values:
- 0 (None)
- 1 (Auto)
- 2 (Fixed)
Default: 1 (Auto)
subfoldername: The name of the folder for
the job processing; it is ignored unless
usesubfolder is set to 2 (Fixed).
report: Generate a report for the job.
createconsolidatedrun: At the end of the job processing, create a new gep file that contains the best models of all the runs in the job, selected according to
different criteria.
Values:
- 0 (Don't Create)
- 1 (Select by best training fitness)
- 2 (select by best validation fitness)
- 3 (select by best training accuracy)
- 4 (select by best validation accuracy)
- 5 (select by best training r-square)
- 6 (select by best validation r-square)
- 7 (select by best training favorite statistic)
- 8 (select by best validation favorite
statistic)
Default: 0 (Don't Create)
async: Process the runs in parallel.
logtofile: Create log files of the run process. Creates one log file per run. The log files are XML files and are named after the run (for example: CreditApproval_000001.xml).
 |
Run Node |
The job node can contain any number of
run nodes. Each run represents a unit of processing which can be processing a run from scratch, continuing the processing of an existing
run or do other tasks defined by other nodes such as code generation and predictions.
The run node is by
far the most complex element in the job file since it can contain so many different nodes and instructions.
Notwithstanding, the simplest run node only needs an id, a stop condition and a type. For example, the run node below attempts to improve the selected model with a run of 50 generations, much like as if you pressed the Continue button in the Run Panel in GeneXproTools.
<run id='1' stopcondition='generations' value='50' type='continue' />
The mandatory run node attributes are:
id: Must be an integer and must be unique. That is, no two run folders can have the same
id.
type: The action to perform with the run. The last four values correspond to the buttons in the Run Panel of GeneXproTools and basically process the run by starting from scratch or by starting from an existing model. Idle is a special case to be used when we need to use the models in the run to generate code or predictions. In this case no new models are created and the attributes
stopcondition and
value are optional.
Values:
- idle
- start
- continue
- simplify
- complexify
stopcondition: Defines the unit for the duration of the run.
Values:
- generations
- hours
- minutes
- seconds
value: The number of generations (integer), hours, minutes or seconds (all float) as defined in
stopcondition to process the run.
 |
Pre and Post Processing (Job and Run) |
The nodes preprocessing and postprocessing can be added inside the job or inside each run. The job's pre and postprocessing nodes are used to start external applications when the job starts and just before the job ends. Similar nodes can be added to each one
of the runs and represent processes that are started just before the run is processed and right after the run ends. The path to the application is defined in the path attribute;
the application can run without showing its window if the
hide attribute is set to yes and the job processor waits for the application to end its processing when the attribute
synchro is set to yes.
path: The fully qualified path to the application to be started.
arguments: A string that is passed to the process being created.
hide: If set to yes then the user interface of the process being fired is hidden.
synchro: Defines if the job processor waits for the process to finish running.
 |
Data Loading |
GeneXproServer can load new training and validation/test datasets into each run or it can completely replace the original data in the run. This operation is attained using the node
dataset which contains a number of attributes and one child node, the
connection. There are five types of
connection: database, file, excel, internal and gepfile.
The dataset node attributes are:
type: Defines the dataset used for
training and validation/testing.
records: The number of records to load. When set to "all" all records in the source are loaded.
Optional Attributes:
reverse: Reverse the dataset (load from last record to the first).
Connection
The connection node determines the type of source data to use. There are five types of connection (database, file, excel, internal and gepfile) that contain information that is specific to each type of data source.
Databases
To load data from a database we use a
connection of
type database as such:
<dataset type="validation" records="500">
<connection type="database" format="responselast">
<oledbconnectionstring>
Provider=Microsoft.Jet.OLEDB.4.0;Data Source=c:\data.mdb;User Id=admin;Password=;"
</oledbconnectionstring>
<sqlstatement>SELECT * FROM Cancer_Test;</sqlstatement>
</connection>
</dataset>
In this example we are selecting the top 500 records of the table Cancer_Test and loading them into the validation dataset.
type: must be a database.
format: The format of the data.
Values:
- responselast: The response (or
dependent) variable is the last column in a tabular dataset.
- responsefirst: The response variable is the first column in a tabular dataset.
- timeseries: The dataset is composed of a single column.
- geneexpression: The dataset in a gene
expression matrix format is transposed.
The database connection contains two nodes:
oledbconnectionstring: A connection string to the database. The connection string must be compatible with ADO (ActiveX Data Objects) technology. The connection string must be set as the value of the node.
sqlstatement: An SQL statement. This is not validated or parsed by GeneXproServer and it is assumed to be correct. It can be of any length and contain any SQL statement that is compatible with the database server in use. The SQL must be set as the value of the node.
Text Files
To load data from a text file you set the
connection type to file as in this example:
<dataset type="training" records="all">
<connection type="file" format="responselast">
<path separator="space" haslabels="no">
c:\Program Files\GeneXproServer 50\samples\1.cancer_train.txt
</path>
</connection>
</dataset>
In this example the file 1.cancer_train.txt contains data separated by spaces and does not have labels in the first row.
type: must be file.
format: The format of the data.
Values:
- responselast: The response (or
dependent) variable is the last column in a tabular dataset.
- responsefirst: The response variable is the first column in a tabular dataset.
- timeseries: The dataset is composed of a single column.
- geneexpression: The dataset in a gene
expression matrix format is transposed.
The file connection node has a single child node called
path that must contain the path to the file as its value. The
path node also requires the following attributes:
separator: The character used to separate the columns.
Values:
- space
- tab
- comma
- semicolon
- pipe
haslabels: Whether the file contains data labels in the first row.
Excel Files
To load data from an Excel file you set the
connection type to excel:
<dataset type='training' records='100'>
<connection type='excel' format='responselast'>
<sheet name='Train_Val' range='$A$1:$P$11' columns='all'>
C:\CreditApproval.xlsx
</sheet>
</connection>
</dataset>
In this example the Excel file CreditApproval.xlsx contains a spreadsheet named Train_Val which contains the data to load in the range $A$1:$P$11. Since the
columns attribute is set to all then all columns will be loaded but since the dataset attribute
records is set to 100 then only the top 100 records will be loaded. It is more efficient to set both these attributes to all and select the exact range.
Excel connection attributes:
type: Must be excel.
format: The format of the data.
Values:
- responselast: The response (or
dependent) variable is the last column in a tabular dataset.
- responsefirst: The response variable is the first column in a tabular dataset.
- timeseries: The dataset is composed of a single column.
- geneexpression: The dataset in a gene
expression matrix format is transposed.
The Excel connection node has a single child node called
sheet that must contain the path to the Excel file as its value. This Excel file must contain at least one spreadsheet with data and the first row of data must be the names of each column (labels).
The sheet node also requires the following attributes:
name: The name of the spreadsheet.
range: The range of columns and rows to load. This range is passed to Excel as is and must be a valid Excel range.
columns: Defines which columns are loaded from the range defined above. To load all columns set
columns to all. To load a subset of the columns this attribute should be set to a list of column names separated by the pipe (|) character. The names of the columns are the values defined in the top row (labels).
GeneXproTools Files
To load data from a gep file you set the
connection type to gepfile:
<dataset type="training">
<connection type="gepfile" gepfiledataset="validation">
<path>C:\BreastCancer.gep</path>
</connection>
</dataset>
In this example we will extract and use all the validation dataset of the BreastCancer.gep file.
GeneXproTools connection attributes:
type: Must be gepfile.
gepfiledataset: The dataset to extract from the gep file.
Values:
- training
- validation
- original
- timeseries
The GeneXproTools connection node has a single child node called
path which contains the path to the file.
Internal Data
The data to load can be part of the job file. In this case it is called internal data as in this example:
<dataset type="training" records="all" reverse="yes">
<connection type="internal" format="responselast">
<data separator="pipe" haslabels="yes">Var0|Var1|Var2|Class
b|22.22|0|1
a|58.67|4.4|0
a|24.5|0.5|1
b|27.83|1.5|1
</data>
</connection>
</dataset>
In this example all the data will be loaded and it is composed of four columns of data separated by
pipes and topped with labels.
Internal data connection attributes:
type: Must be
internal.
format: The format of the data.
Values:
- responselast: The response (or
dependent) variable is the last column in a tabular dataset.
- responsefirst: The response variable is the first column in a tabular dataset.
- timeseries: The dataset is composed of a single column.
- geneexpression: The dataset in a gene
expression matrix format is transposed.
The internal data connection must contain a
data node with the data itself as its value and the following attribute set:
separator: The character used to separate the columns.
Values:
- space
- tab
- comma
- semicolon
- pipe
haslabels: Whether the data table contains labels in the first row.
 |
Changing Settings |
When a job processes a run, it uses all the settings in the original run.
However it is possible to adjust the settings of each individual run using the
settings node and its child the
setting node.
The settings node can contain any number of
setting nodes which specify the setting and the value to change.
For example:
<settings>
<setting key='Genes' value='5'/>
<setting key='HeadSize' value='10'/>
</settings>
In this example we are changing the Genes value to 5 and the HeadSize to 10. The keys and values correspond to the settings in GeneXproTools with the same name. These values are not validated so you must ensure that the correct type is used or the run file may become corrupted. The list of settings that can be changed is shown in the settings page.
Some settings change the structure of the models and in this case GeneXproServer gives a warning and clears all the models in the run. The settings in question are:
- HeadSize
- Genes
- LinkingFunction
- TimeSeriesDelayTime
- TimeSeriesEmbeddingDimension
- UseRNC
- ConstantsPerGene
 |
Changing the Function Set |
The functions node allows adding and removing functions as well as changing the weight of the functions that are part of the function set:
The functions node can contain any number of
function nodes which specify the
action and the
symbol to change. See the example:
<functions>
<function action="add" symbol="Pow" weight="2"/>
<function action="set" symbol="+" weight="5"/>
<function action="remove" symbol="*"/>
</functions>
In this example we are adding the function Pow (power) to the function set with a weight of 2; we are increasing the weight of the
addition (+) function to 5; and, finally, we are removing the
multiplication (*) function from the function set.
When you are continuing a run (Continue, Simplify and Complexify run options) you must ensure that you are not removing a function that is used in a model of the original run and that you are not adding a function with higher arity than the maximum arity of the run. If you are starting a run from scratch then the only limitation is that you should not remove all the functions from the function set.
Each function node must contain the following three attributes:
action: whether to add, remove or change the weight of the function.
Values:
- add: Adds the function to the function set.
- remove: Removes the function from the function set.
- set: Changes the weight of the function.
symbol: the representation of the function (see
the Functions entry for a list of supported symbols).
weight: the weight of the function in the function set (integer).
If you add a function which has higher arity than the max arity of the current function set, then GeneXproServer gives a warning and clears all the models in the run. The same happens if the removal of a function causes the max arity of the function set to change.
 |
Time Series Prediction Modes |
In GeneXproServer and GeneXproTools the Time Series
Prediction category can run in two prediction modes: Testing and Prediction. It is also possible to
go back and forward between prediction modes and to do this operation in GeneXproServer we use the
transform node. This node has one attribute that determines which
prediction mode the run is being converted to:
<job filename='Sunspots.gep' path='c:\data' feedback='2' >
<run id='1' type='idle'>
<transform name='timeseriestopredictionmode'/>
</run>
</job>
In this example the prediction mode of the run Sunspots.gep will be changed to
Prediction. If the run is already in prediction mode then nothing
is changed.
The following example performs the opposite operation, changing the
prediction mode to testing, and also modifying the number of
testing predictions (optional):
<job filename='Sunspots.gep' path='c:\data' feedback='2' >
<run id='1' type='idle'>
<transform name='timeseriestotestingmode'/>
<settings>
<setting key='TimeSeriesTestingPredictions' value='5'/>
</settings>
</run>
</job>
name: The type of transformation.
Values:
- timeseriestopredictionmode: Change the
prediction mode to Prediction.
- timeseriestotestingmode: Change the
prediction mode to Testing.
 |
Setting the Singled Out Class |
The Classification and Logistic Regression categories support datasets with
multiple classes in the response variable. With GeneXproServer
it is possible to change which of these classes is assigned the value 1 (all the other classes are assigned the value 0). This transformation is applied before processing the run and
uses the transform node:
<job filename='IrisPlants.gep' path='c:\data' feedback='2' >
<run id='1' type='idle'>
<transform name='setsingledoutclass'>IrisSetosa</transform>
</run>
</job>
The example above sets the class IrisSetosa to be the singled out class. Note that the name of the singled out class must exactly match one of the classes in the response variable or the transformation will fail.
 |
Testing |
At the end of a run with a validation/test dataset, GeneXproServer tests
just the last model on the validation/test dataset by default;
all the other intermediate models are left untested by
default. If you want to test all the models or a number of models
against the validation/test dataset, then you can use the
test node:
<job filename='CreditApproval.gep' path='c:\work'
feedback='2' usesubfolder='1' async='yes'>
<run id='1' stopcondition='Minutes' value='30'>
<test whichmodels='all'/>
</run>
</job>
In this example the run will be processed for 30 minutes and, at the end
of the run, it will test all the models on the validation
dataset. If instead of all
the value of the attribute was 10, for example, it would
test the last 10 models only:
<job filename='CreditApproval.gep' path='c:\work'
feedback='2' usesubfolder='1' async='yes'>
<run id='1' stopcondition='Minutes' value='30'>
<test whichmodels='10'/>
</run>
</job>
Finally, you can specify the dataset to test by adding a
dataset attribute which can have the values
training,
validation or
both. The first will test the training set, the second, which is the default
and can be omitted, will test the validation set and
both will test both datasets. This can be very useful if you change the fitness function
or the favorite statistic and want to recalculate their values for a large number of models. This functionality is similar to the Refresh/Test buttons in the History Panel of GeneXproTools.
<job filename='CreditApproval.gep' path='c:\work'
feedback='2' usesubfolder='1' async='yes'>
<run id='1' stopcondition='Minutes' value='30'>
<test whichmodels='all' dataset='both'/>
</run>
</job>
In this case all the models will be tested against the training and validation datasets.
whichmodels: either
all or a positive integer.
dataset:
defines the dataset to be tested.
 |
Pre-Selecting and Selecting Models |
There are two selection nodes: the
preselect and the
select nodes. Both set a model to be the active or current model according to a number of criteria or to a model index. The
preselect operation sets the model to active before the run starts and it is useful when you want to continue a run from a model that was not
the active model in the original run. The
select node changes the active model at the end of a run and can be useful when generating code
from models that are not the last one.
<run id='1' type='continue' stopcondition='generations' value='500'>
<!-- Select model number 3 before starting the run -->
<preselect criteria='modelindex' value='3'/>
<!-- Select model with best favorite statistic at the end of the run -->
<select criteria='bestvalidationfitness'/>
</run>
<run id='2' type='continue' stopcondition='generations' value='500'>
<settings>
<setting key='FavoriteStatistic' value='36'/>
</settings>
<test whichmodels='all' dataset='both'/>
<!-- Select model with best average training and validation fitness before starting the run -->
<preselect criteria='avgtrainingvalidationfitness'/>
<!-- Select model with best average training and validation favorite statistic at the end of the run -->
<select criteria='avgtrainingvalidationfavorite'/>
</run>
In the first run of this example GeneXproServer selects model number 3, then improves that model for 500 generations, after which it sets the model with the best
fitness value in the validation dataset to be the active model. When the
select node is not specified GeneXproServer always selects the model with the best training fitness.
In the second run of this example GeneXproServer selects
the model with the best average fitness value in the
training and validation datasets to be used as seed
model in another optimization run of 500 generations.
Then at the end of the run it selects the model with the
best average value for the favorite statistic in the
training and validation datasets. The extra code for
setting the favorite statistic (the area under the ROC
curve in this case) and testing all models on
the training and validation datasets must be included if
you want to apply any of the selection criteria that
involve favorite statistics.
criteria: The type of selection.
Values:
- besttrainingfitness
- bestvalidationfitness
- lastmodel
- firstmodel
- random
- besttrainingfavorite
- bestvalidationfavorite
- avgtrainingvalidationfavorite
- avgtrainingvalidationfitness
- modelindex
value: Integer that matches the model
index in a run. Only used when criteria is set to modelindex.
 |
Converting Model to Code |
GeneXproServer lets you convert any model to the 19 supported programming languages or to your own custom programming language. For this end it uses the
convert node. This node has several attributes that give you control over the language, the format of the code,
the output type for CLassification and Logistic
Regression runs and the location where it is saved.
<run id='1' type='continue' stopcondition='generations' value='500'>
<convert language="javascript" uselabels="yes" format="text"
filename="MyModel" outputtype="MostLikelyClass" grammartype=""/>
</run>
This examples converts the last model to Javascript and saves it to the file
MyModel.js (filename) next to the run. The code will use the labels for the variable names (uselabels) and
will output the most likely class (outputtype).
language: Any of the 19 supported
programming languages or the name of a custom language.
uselabels: When set to yes the code uses the variable labels for the variable names in the model. Otherwise it uses the
default d0, d1,…dn representation.
format: The format of the model. This allows exporting the model for external systems (xml) or to serve to web clients directly (json).
filename: The name of the file where the converted model will be saved. It should not have an extension since GeneXproServer will add the programming language extension to the end.
outputtype: This attribute corresponds to the
Model Output Types used in GeneXproTools Classification and Logistic Regression runs.
Values:
- rawmodel
- mostlikelyclass
- probability1
grammartype: This is only applicable in Logistic Synthesis runs and corresponds to the type of gates used to build the model's code.
Values:
- allgates
- notandoronly
- nandonly
- noronly
- muxsystem
- reedmullersystem
 |
Consolidated Runs |
Very long runs can produce a very large number of models
that need to be analyzed at the end of a job. To help
with model selection GeneXproServer can pick a
particular model from each run in a job according to a
certain criterion and add them all to a new run file.
Usually you would select the best model of each run
according to the selection criterion you are interested in.
<job filename='BreastCancer.gep' path='C:\data' createconsolidatedrun='4'>
<run id='1' stopcondition='Minutes' value='30'/>
<run id='2' stopcondition='Minutes' value='45'/>
<run id='3' stopcondition='Minutes' value='45'/>
<run id='4' stopcondition='Minutes' value='45'/>
<run id='5' stopcondition='Minutes' value='45'/>
<run id='6' stopcondition='Minutes' value='45'/>
<run id='7' stopcondition='Minutes' value='45'/>
</job>
In this example we have 7 runs that will run for some time each and at the end GeneXproServer will create a file named BreastCancer_consolidated.gep with 7 models which were selected from
each of the generated runs because they had the best accuracy in the validation set.
createconsolidatedrun: Create a new run file with models selected from the generated runs in the job.
Values:
- 0: Don't Create
- 1: Select based on best Training Fitness
- 2: Select based on best Validation Fitness
- 3: Select based on best Training Accuracy
- 4: Select based on best Validation Accuracy
- 5: Select based on best Training R-square
- 6: Select based on best Validation R-square
- 7: Select based on best Training Favorite Statistic
- 8: Select based on best Validation Favorite Statistic
 |
Making Predictions (Time Series Prediction) |
GeneXproServer lets you automate the creation of predictions and can export them to a number of formats. This can be achieved using xml similar to the example below:
<job filename=Dowjones.gep' path='C:\data'>
<run id='1' stopcondition='Minutes' value='0.3'>
<predict quantity='3' format='xml' filename='auto'/>
</run>
</job>
In this example the run is processed for 0.3 minutes and then 3 predictions are generated using the last model. These predictions are saved in xml format to a file that is derived from the run’s filename (DowJones_000001_predictions.xml in this case).
quantity: The number of predictions to generate (positive integer).
format: The format used to lay the predictions in the file.
filename: Either a filename or the word
auto. When set to
auto the predictions file will be named after the run name with the posfix _predictions.
 |
Sync/Async Processing |
The current version of GeneXproServer introduces a very efficient parallel processing algorithm that allows it to scale linearly to
a large number of cores. By default all runs are now processed in parallel but you can revert to single run processing by setting the
async attribute of the job node to
no. This may be important when you are using external fitness functions that cannot service more than one run at a time.
Each run is processed in a different Windows process which implies that they are completely isolated from each other. GeneXproServer queues all the runs at the beginning of a job but only processes as many as the number of cores available simultaneously.
<job filename='BreastCancer.gep' async='yes' path='C:\runs'>
<run id='1' stopcondition='Minutes' value='30'/>
<run id='2' stopcondition='Minutes' value='45'/>
<run id='3' stopcondition='Minutes' value='45'/>
<run id='4' stopcondition='Minutes' value='45'/>
<run id='5' stopcondition='Minutes' value='45'/>
<run id='6' stopcondition='Minutes' value='45'/>
<run id='7' stopcondition='Minutes' value='45'/>
</job>
In this example, which assumes a 4 core CPU and that all runs are equal in structure and dataset, GeneXproServer will process the first
4 runs simultaneously until the minute 30 where run number 1 finishes and GeneXproServer starts run 5. At
minute 45 the runs 2 to 4 will end and the runs 6 and 7 are started. Run 5 will end 30 minutes later and the last two will run for a further 15 minutes. These values will vary due to the
random nature of the algorithms but the total processing time will be close to a fourth of the time it would take to process these runs serially.
|

